ABOUT RIVER
Content_dbRIVER_202411.sql.gz contains 5 tables, with the schema outlined below:
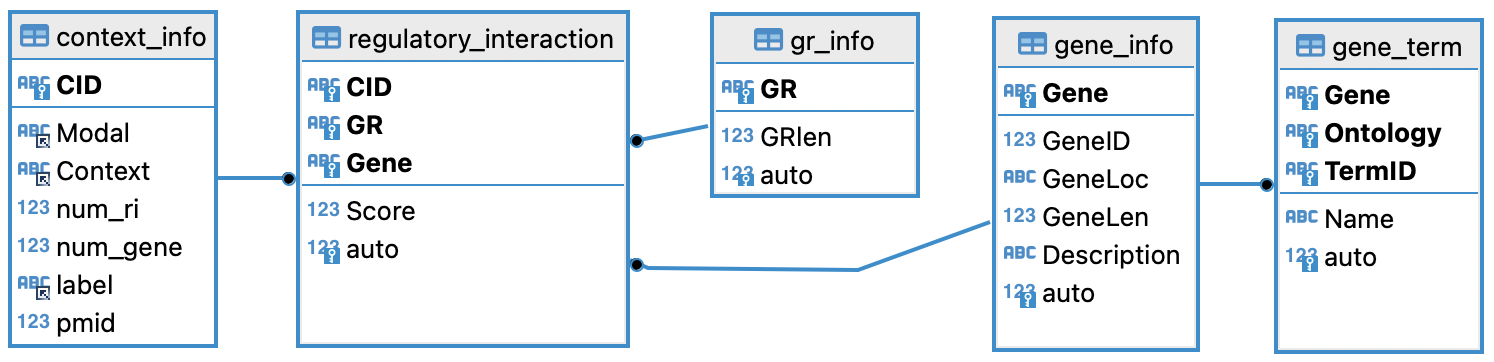
|
|
|
USE
Restore the RIVER MySQL database
# Enter password if prompted
mysql -u root -p -e "create database RIVERdb;"
gunzip < Content_dbRIVER_202411.sql.gz | mysql -u root -p dbRIVER
Show tables
mysql> USE dbRIVER; mysql> SHOW tables; +------------------------+ | Tables_in_dbriver | +------------------------+ | context_info | | gene_info | | gene_term | | gr_info | | regulatory_interaction | +------------------------+
Extract genomic regions that interact with the gene 'IL17A' (based on the modality 'PCHiC')
mysql> SELECT a.CID, b.label, a.GR, a.Gene, a.Score FROM regulatory_interaction AS a, context_info AS b WHERE a.CID=b.CID AND b.Modal='PCHiC' AND a.Gene='IL17A' ORDER BY a.Score DESC; +-------+-----------------------------------------------+------------------------+-------+--------------+ | CID | label | GR | Gene | Score | +-------+-----------------------------------------------+------------------------+-------+--------------+ | C0081 | Aorta (PMID31501517) | chr6:50715234-50721151 | IL17A | 0.00886063 | | C0088 | Fat (PMID31501517) | chr6:52021964-52028582 | IL17A | 0.00793775 | | C0080 | AdrenalGland (PMID31501517) | chr6:50138944-50145136 | IL17A | 0.00741363 | | C0126 | hippocampal (PMID31367015) | chr6:52107437-52120696 | IL17A | 0.00652897 | | C0123 | Trophoblast (PMID31501517) | chr6:52121367-52124792 | IL17A | 0.00572892 | | C0112 | Pancreas (PMID31501517) | chr6:52028583-52033567 | IL17A | 0.00566848 | | C0123 | Trophoblast (PMID31501517) | chr6:52874476-52880638 | IL17A | 0.00491858 | | C0081 | Aorta (PMID31501517) | chr6:51983487-51987113 | IL17A | 0.00432594 | | C0112 | Pancreas (PMID31501517) | chr6:51976923-51980357 | IL17A | 0.00291745 | | C0110 | Nonactivated_total_CD4_T_cells (PMID27863249) | chr6:52507448-52510680 | IL17A | 0.00215937 | | C0082 | Bladder (PMID31501517) | chr6:52097457-52102314 | IL17A | 0.00208858 | | C0123 | Trophoblast (PMID31501517) | chr6:51930792-51937411 | IL17A | 0.00140648 | | C0107 | Naive_CD4_T_cells (PMID27863249) | chr6:52516121-52519136 | IL17A | 0.00116485 | | C0110 | Nonactivated_total_CD4_T_cells (PMID27863249) | chr6:52548793-52551154 | IL17A | 0.00109584 | | C0097 | LCL (PMID31501517) | chr6:50706937-50711709 | IL17A | 0.00102056 | | C0079 | Activated_total_CD4_T_cells (PMID27863249) | chr6:52527279-52528288 | IL17A | 0.000919697 | | C0111 | Ovary (PMID31501517) | chr6:52097457-52102314 | IL17A | 0.000908583 | | C0108 | Naive_CD8_T_cells (PMID27863249) | chr6:52516121-52519136 | IL17A | 0.000851565 | | C0122 | Total_CD8_T_cells (PMID27863249) | chr6:52516121-52519136 | IL17A | 0.000743387 | | C0079 | Activated_total_CD4_T_cells (PMID27863249) | chr6:52564858-52565095 | IL17A | 0.000726709 | | C0121 | Total_CD4_T_cells (PMID27863249) | chr6:52529945-52533082 | IL17A | 0.000650316 | | C0079 | Activated_total_CD4_T_cells (PMID27863249) | chr6:52565096-52568603 | IL17A | 0.000545248 | | C0109 | Neutrophils (PMID27863249) | chr6:50770604-50780906 | IL17A | 0.000543862 | | C0109 | Neutrophils (PMID27863249) | chr6:50796768-50800786 | IL17A | 0.000511279 | | C0079 | Activated_total_CD4_T_cells (PMID27863249) | chr6:52483390-52486722 | IL17A | 0.000504013 | | C0123 | Trophoblast (PMID31501517) | chr6:51998371-52001727 | IL17A | 0.000475964 | | C0122 | Total_CD8_T_cells (PMID27863249) | chr6:52582960-52583875 | IL17A | 0.000459409 | | C0123 | Trophoblast (PMID31501517) | chr6:52168413-52176918 | IL17A | 0.000450113 | | C0099 | Liver (PMID31501517) | chr6:51365528-51370740 | IL17A | 0.000427498 | | C0108 | Naive_CD8_T_cells (PMID27863249) | chr6:52564858-52565095 | IL17A | 0.000414165 | | C0108 | Naive_CD8_T_cells (PMID27863249) | chr6:52551155-52557252 | IL17A | 0.000373828 | | C0107 | Naive_CD4_T_cells (PMID27863249) | chr6:52507448-52510680 | IL17A | 0.00034523 | | C0110 | Nonactivated_total_CD4_T_cells (PMID27863249) | chr6:52557253-52561729 | IL17A | 0.000311453 | | C0110 | Nonactivated_total_CD4_T_cells (PMID27863249) | chr6:52564858-52565095 | IL17A | 0.000303343 | | C0121 | Total_CD4_T_cells (PMID27863249) | chr6:52582960-52583875 | IL17A | 0.000257019 | | C0122 | Total_CD8_T_cells (PMID27863249) | chr6:52506678-52507447 | IL17A | 0.000244341 | | C0109 | Neutrophils (PMID27863249) | chr6:50766439-50770603 | IL17A | 0.000182812 | | C0121 | Total_CD4_T_cells (PMID27863249) | chr6:52551155-52557252 | IL17A | 0.000175933 | | C0127 | motor (PMID31367015) | chr6:52120697-52122033 | IL17A | 0.000166591 | | C0110 | Nonactivated_total_CD4_T_cells (PMID27863249) | chr6:52583876-52584149 | IL17A | 0.000141738 | | C0107 | Naive_CD4_T_cells (PMID27863249) | chr6:52582960-52583875 | IL17A | 0.000110967 | | C0122 | Total_CD8_T_cells (PMID27863249) | chr6:52474654-52478765 | IL17A | 0.0000768145 | | C0079 | Activated_total_CD4_T_cells (PMID27863249) | chr6:52608673-52609968 | IL17A | 0.0000613927 | | C0110 | Nonactivated_total_CD4_T_cells (PMID27863249) | chr6:52537239-52538716 | IL17A | 0.0000469573 | | C0110 | Nonactivated_total_CD4_T_cells (PMID27863249) | chr6:52565096-52568603 | IL17A | 0.0000382299 | | C0121 | Total_CD4_T_cells (PMID27863249) | chr6:52507448-52510680 | IL17A | 0.000033339 | +-------+-----------------------------------------------+------------------------+-------+--------------+
context_info
A table about contexts Table_context_info.txt.gz
mysql> DESC context_info;
+----------+---------------------------+------+-----+---------+-------+
| Field | Type | Null | Key | Default | Extra |
+----------+---------------------------+------+-----+---------+-------+
| CID | varchar(5) | NO | PRI | NULL | |
| Modal | enum('ABC','PCHiC','QTL') | NO | MUL | NULL | |
| Context | varchar(100) | NO | MUL | NULL | |
| num_ri | int unsigned | NO | | 0 | |
| num_gene | int unsigned | NO | | 0 | |
| label | varchar(100) | NO | MUL | NULL | |
| pmid | int unsigned | NO | | 0 | |
+----------+---------------------------+------+-----+---------+-------+
- The CID column is the context ID
- The Modal column is the modality for regulatory interactions
- The Context column is the name of contexts
- The num_ri column is the number of regulatory interactions
- The num_gene column is the number of genes
- The label column is the human-friendly description of contexts
- The pmid column is PubMed ID for this published dataset
regulatory_interaction
A table about regulatory interactions Table_regulatory_interaction.txt.gz
mysql> DESC ri;
+-------+--------------+------+-----+---------+----------------+
| Field | Type | Null | Key | Default | Extra |
+-------+--------------+------+-----+---------+----------------+
| CID | varchar(5) | NO | PRI | NULL | |
| GR | varchar(50) | NO | PRI | NULL | |
| Gene | varchar(100) | NO | PRI | NULL | |
| Score | double | YES | | NULL | |
| auto | int | NO | UNI | NULL | auto_increment |
+-------+--------------+------+-----+---------+----------------+
- The CID column is the context ID
- The GR column is the genomic region
- The Gene column is the official gene symbol
- The Score column is the score for regulatory interactions
- The auto column is an increasing sequence of unique numbers, collectively acting as a primary key
gr_info
A table about genomic regions Table_gr_info.txt.gz
mysql> DESC gr_info;
+-------+--------------+------+-----+---------+----------------+
| Field | Type | Null | Key | Default | Extra |
+-------+--------------+------+-----+---------+----------------+
| GR | varchar(50) | NO | PRI | NULL | |
| GRlen | int unsigned | NO | | 0 | |
| auto | int | NO | UNI | NULL | auto_increment |
+-------+--------------+------+-----+---------+----------------+
- The GR column is the genomic region
- The GRlen column is the genomic region length (calculated based on GR)
- The auto column is an increasing sequence of unique numbers, collectively acting as a primary key
gene_info
A table about human genes Table_gene_info.txt.gz
mysql> DESC gene_info;
+-------------+--------------+------+-----+---------+----------------+
| Field | Type | Null | Key | Default | Extra |
+-------------+--------------+------+-----+---------+----------------+
| Gene | varchar(100) | NO | PRI | NULL | |
| GeneID | int unsigned | NO | | 0 | |
| GeneLoc | varchar(50) | NO | | NULL | |
| GeneLen | int unsigned | NO | | 0 | |
| Description | varchar(255) | NO | | NULL | |
| auto | int | NO | UNI | NULL | auto_increment |
+-------------+--------------+------+-----+---------+----------------+
- The Gene column is the official gene symbol
- The GeneID column is the NCBI Gene ID
- The GeneLoc column is the genomic location (derived from UCSC known genes)
- The GeneLen column is the gene length (calculated based on GeneLoc)
- The Description column is the official gene description
- The auto column is an increasing sequence of unique numbers, collectively acting as a primary key
gene_term
A table about gene annotations using ontology terms Table_gene_term.txt.gz
mysql> DESC gene_info;
+----------+--------------+------+-----+---------+----------------+
| Field | Type | Null | Key | Default | Extra |
+----------+--------------+------+-----+---------+----------------+
| Gene | varchar(100) | NO | PRI | NULL | |
| Ontology | char(8) | NO | PRI | NULL | |
| TermID | varchar(50) | NO | PRI | NULL | |
| Name | varchar(255) | NO | | NULL | |
| auto | int | NO | UNI | NULL | auto_increment |
+----------+--------------+------+-----+---------+----------------+
- The Gene column is the official gene symbol
- The Ontology column is the Ontology
- The TermID column is the ontology term ID
- The Name column is the ontology term name
- The auto column is an increasing sequence of unique numbers, collectively acting as a primary key